Run Jupyter notebooks quietly from command-line#
runPyNB is a quick and dirty utility to run (and time) Jupyter notebooks from command-line and makefiles.
Quickstart#
Install from PyPI
pip install runpynb
General usage: runpynb <notebook(s)> [options] (“.ipynb” not required)
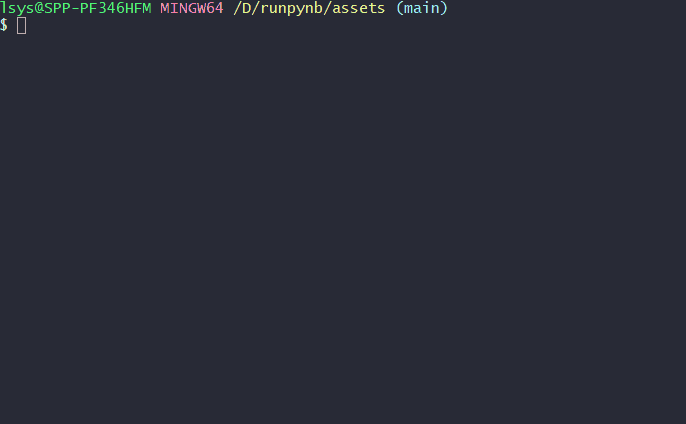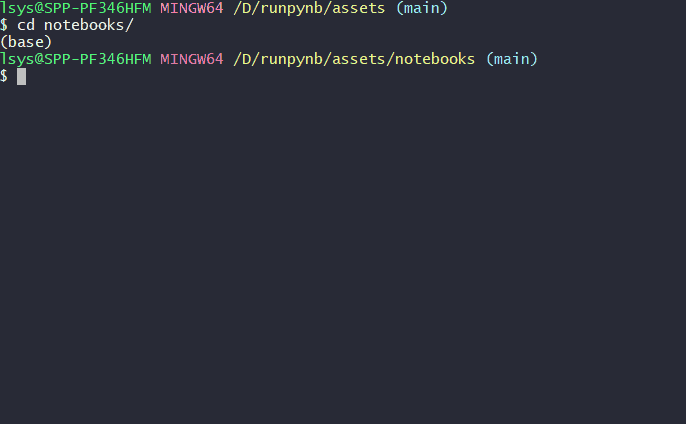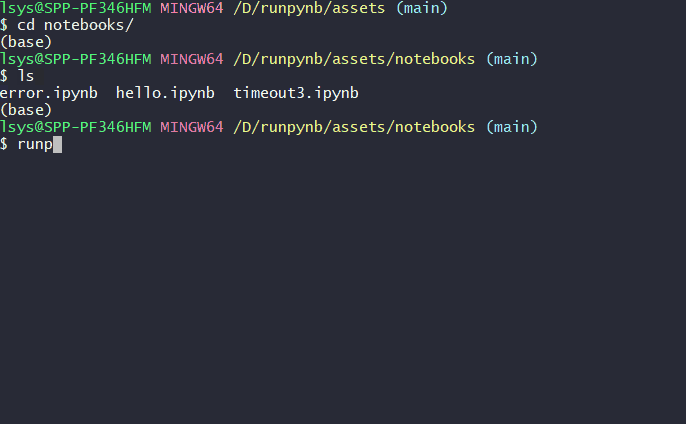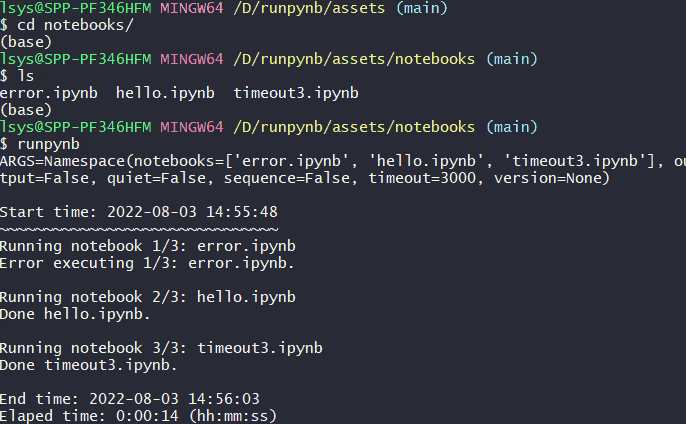
runpynb: Run all notebooks in directory.$ runpynb
Usage#
runpynb <notebook(s)> -q: Run quietly (-q).$ runpynb hello.ipynb -q

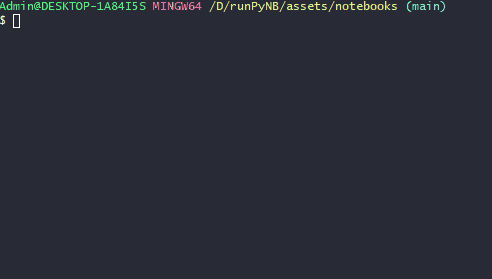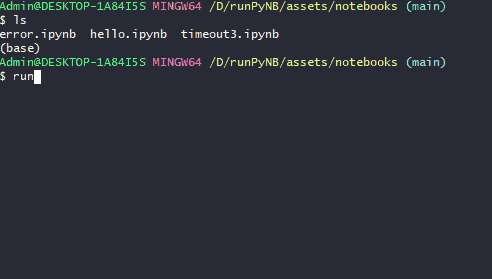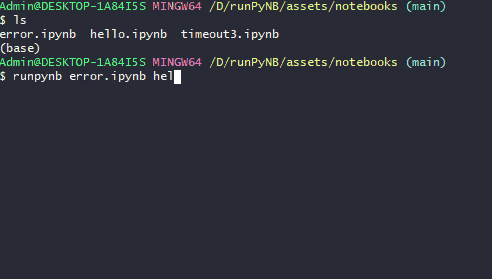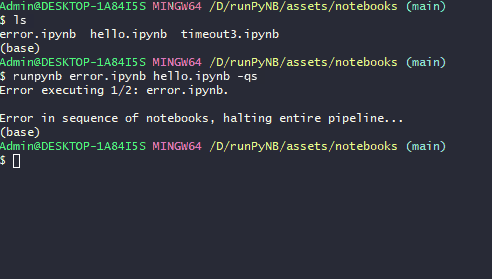
runpynb <notebook(s)> -qs: Run quietly (-q) as a sequence of workflow (-s). Errors (eg inerror.ipynb) will break the workflow.$ runpynb error.ipynb hello.ipynb -qs
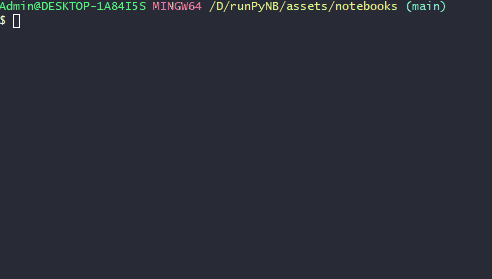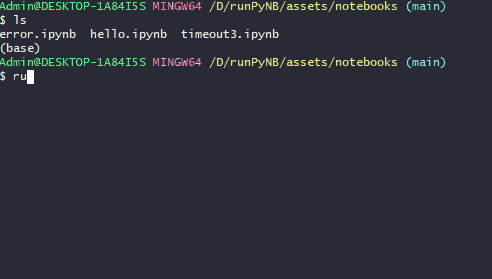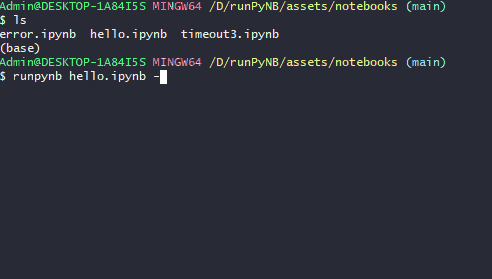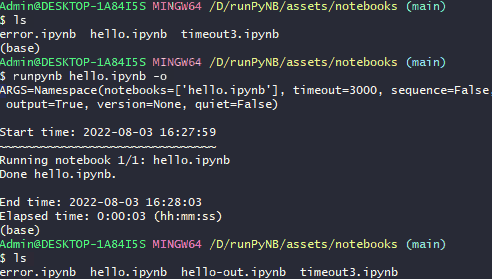
runpynb <notebook(s)> -o: Save output as separate notebook (-o), instead of overwriting existing notebook(s).$ runpynb hello.ipynb -o
Options#
usage: runpynb [-h] [-t TIMEOUT] [-s] [-o] [-v VERSION] [-q] [notebooks ...]
Run (and time) Jupyter notebooks silently in command-line.
positional arguments:
notebooks List of Jupyter notebooks (*.ipynb) to be run
(default=all notebooks in path).
optional arguments:
-h, --help show this help message and exit
-t TIMEOUT, --timeout TIMEOUT
Seconds until a cell in the notebook timesout, which
raises a Timeouterror exception (default is 3000=5
mins).
-s, --sequence Sequence implicit in notebook lists. If error occurs
somewhere, stop entire pipeline.
-o, --output Save output as a separate notebook with "-out"-suffix
(e.g. *-out.ipynb) instead of overwriting existing
file.
-v VERSION, --version VERSION
Version of notebook to return (Default=No conversion).
Notebook will be converted if necessary.
-q, --quiet Be quiet and don't print messages (including run
time). Caution: Does not suppress error messages.
Status#
More on this package#
This is a lightweight package that wraps around the official Jupyter nbformat and nbconvert modules.
My workflow involves using Jupyter notebooks to clean, and analyze data.
I use this utility to run notebooks silently from the command-line and Makefiles (without converting from .ipynb files to .py files).
Related packages are guoquan/runnb and vinayak-mehta/nbcommands with a planned enhancement nbtime to run Jupyter notebooks from command-line.
Usage with Makefiles#
A minimal workflow where get-data.ipynb takes 5000 seconds to prepare data.csv.
And where analyze.ipynb uses data.csv to produce output.png.
.DEFAULT_GOAL := output.png
data.csv: get-data.ipynb
runpynb $^ -t 5000
output.png: analyze.ipynb data.csv
runpynb $<
Known Issues#
Build fails with Python 3.6 in Windows OS.
Notebooks with long execution time will require the
timeoutoption (egrunpynb notebook.ipynb -t 10000).
License#
This package is licensed under the MIT License.


